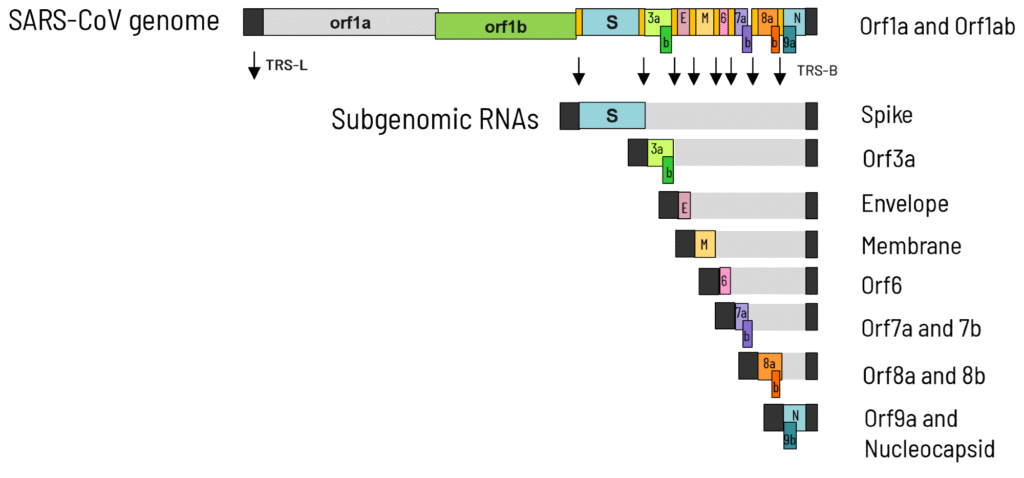
At the ELRIG Drug Discovery 2025 conference, researchers from IRBM S.p.A. presented a poster detailing their advancements in developing antiviral strategies for coronavirus infections. The team, composed of various authors including Cristina Alli and Chiara Soldati, introduced a stable cellular model for studying SARS-CoV replication using an RNA replicon. This replicon lacks structural protein coding sequences but is engineered to express NanoLuc Luciferase, functioning as a reporter gene.
They focused on the discontinuous transcription mechanism, which is essential for producing subgenomic mRNAs necessary for viral replication. The model enables high-throughput screening (HTS) of a compound library containing approximately 413,000 entries. Following extensive evaluation, they optimized assay conditions to monitor antiviral activity effectively. The study identified 3,371 potential hits, leading to 2,232 candidates for further potency and cytotoxicity testing.
Currently, the research team is examining the interference of selected compounds with the assay readout and their effects on unrelated RNA viruses. This research aims to enhance understanding of coronavirus replication and to identify viable therapeutic inhibitors.
- Why it matters: This research could lead to the development of effective treatments for coronavirus infections, potentially impacting public health and pharmaceutical strategies.
- The latest: The compound library screening revealed 3,371 hits, with follow-up assays currently underway for further characterization.
Source: https://www.news-medical.net/health/Identifying-coronavirus-transcription-inhibitors-through-phenotypic-screening.aspx
Source: https://www.news-medical.net/health/Identifying-coronavirus-transcription-inhibitors-through-phenotypic-screening.aspx